Configuration file
Note
The configuration files allow to set the integrated studies (coarsening and history matching) only for the drogon and norne model. To use pycopm in any given OPM Flow geological model to generate modified files (coarsening, refinement, submodels, and transformations), this can be achieved without a configuration file, but setting the parameters via command lines (see the Overview or run pycopm -h for the definition of the argument options, as well as the examples in Via OPM Flow decks.)
Here we use as an example one of the configuration files used in the tests (see input.toml). The first input parameter is:
1# Set mpirun, the full path to the flow executable, and simulator flags (except --output-dir)
2flow = "flow --newton-min-iterations=1"
If flow is not in your path, then write the full path to the executable, as well as adding mpirun if this is supported in your machine (e.g., flow = “mpirun -np 8 /Users/dmar/Github/opm/build/opm-simulators/bin/flow --newton-min-iterations=1”).
The next entries define the following parameters:
4# Set the model parameters
5field = "norne" # Geological model (norne or drogon)
6mode = "ert" # Mode to run (single-run, files, or ert)
7X = [0,2,0,2,2,0,2,0,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,0,2,0,2,2,0,2,2,0,2,2,2,2,0] # Array of x-coarsening
8Y = [0,2,0,2,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,2,0,2,0,2,0,2,0,2,0,2,0,2,0,2,0,2,0,2,0,2,2,2,2,2,2,2,2,2,0] # Array of y-coarsening
9Z = [0,0,2,0,0,2,2,2,2,2,0,2,2,2,2,2,0,0,2,0,2,2,0] # Array of z-coarsening
10net = 2 # Number of ensembles
11mep = 2 # Maximum number of ensembles running in parallel
12mrt = 600 # Maximum runtime in seconds of a realization. A value of 0 means unlimited runtime.
13mrn = 2 # Minimum number of realizations that must have succeeded for the simulation to be regarded as a success.
14rds = 7 # Set a specific seed for reproducibility. A value of 0 means no seed.
15obs = "observations_training" # Name of the observation file for the hm ('observations_training', 'observations_test', or 'observations_complete')
16deck = 0 # Select which coarser deck to use: 0 -> default one or 1 -> LET sat functions
17letsatn = 1 # For norne: for the LET coarser deck, select: 0 -> SATNUM=1, 1 -> SATNUM is computed from Sandve et al 2022, 2 -> #SATNUM=#Cells.
18cporv = 0 # 0 -> no corrections for lost porv, 1 -> correct it on the cell boundaries, 2 -> account it on the porosity on all cells
19initial = 0 # Initialization 0 -> Equil 1->INIT from fine-scale
20error = [0.1,0.1,0.1] # Error WWPR, WOPR, and WGPR
21minerror = [100,100,100000] # Minimum error of WWPR, WOPR, and WGPR
22date = 2005-03-01 # Last date to HM
23suffixes = ["PRT"] # Delete files with this suffix to save storage
The single-run mode results in Flow running only one simulation and the information shown in the terminal is the one from the Flow executable instead of the ERT one. The ert option calls the ERT executable and the command values are given after in the same input file which are discussed later. The files option only writes the needed input files for ERT.
The X, Y, and Z inputs define which pillars are removed (value 2) in the x, y, and z direction respectively.
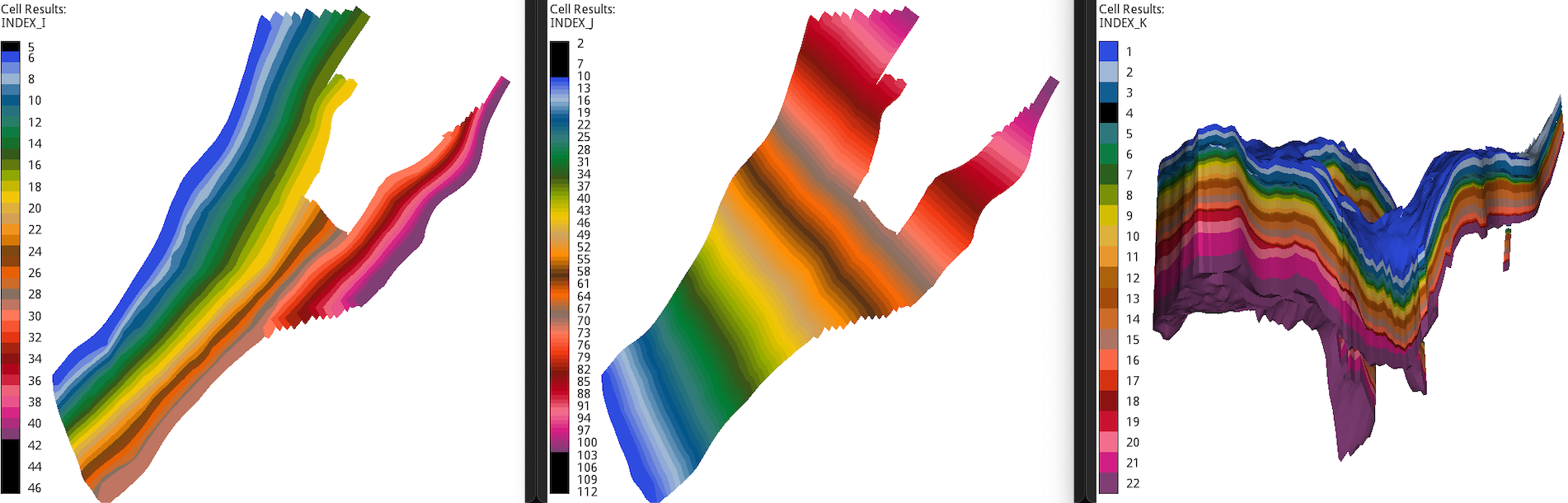
I, J, and K cell index in the standard Norne data set.
Values of 0 do not remove the pilar.
Tip
The 0 values are used to keep pillars in order to honor the main Norne shape. Then from the example provided here, change the 2’s to 0’s to add back the pillars.
The remaining lines define the number of ensembles, maximum number of ensembles running in parallel, the maximum runtime in seconds to stop a realization (0 means unlimited time), the minimum number of realizations for the simulation to be regarded as a success, the random seed for reproducibility (0 means no seed), the observation file for the hm (‘observations_training.data’, ‘observations_test.data’, or ‘observations_complete.data’), to use the saturation functions from opm-tests norne or to use the LET saturation functions, defining if each of the coarser cells is considered as only one region, regions as in Sandve et al 2022 or as a different SATNUM region, and the cporv entry sets if the ntg and poro properties on the cells on the boundary are modified to account for lost PV respect to the default simulation or if the porosity of all cells is modified to match this value. The last entry set the type of files to delete after each realization is completed to save memory.
If the ert option is activated, then the following input:
25# Set the command line for the ert executable (gui, test_run, ensemble_experiment, ensemble_smoother, iterative_ensemble_smoother, and es_mda) and flags
26ert = "es_mda --weights 1"
sets the type of ert option and command flags. Currently, the options supported are gui, test_run, ensemble_experiment, ensemble_smoother, iterative_ensemble_smoother, and es_mda. Confer to the ERT documentation for a full description of these options.
The LET saturation function parameters for each of the coarsened cells are given in the following entry:
28# Properties LET saturation functions: name, value, use dist in hm?, dist, distpara, distpara
29LET = [["lw", 3, 1, "UNIFORM", 1.1, 5],
30["ew", 1, 1, "UNIFORM", -1, 2],
31["tw", 3, 1, "UNIFORM", 1.1, 5],
32["lo", 3, 1, "UNIFORM", 1.1, 5],
33["eo", 1, 1, "UNIFORM", -1, 2],
34["to", 3, 1, "UNIFORM", 1.1, 5],
35["lg", 3, 1, "UNIFORM", 1.1, 5],
36["eg", 1, 1, "UNIFORM", -1, 2],
37["tg", 3, 1, "UNIFORM", 1.1, 5],
38["log", 3, 1, "UNIFORM", 1.1, 5],
39["eog", 1, 1, "UNIFORM", -1, 2],
40["tog", 1, 1, "UNIFORM", 1.1, 5],
41["lmlto", 1.5, 1, "UNIFORM", 1, 2],
42["emlto", 1, 1, "UNIFORM", 0.9, 2.1],
43["tmlto", 1.5, 1, "UNIFORM", 1, 2],
44["lmltg", 1.5, 1, "UNIFORM", 1, 2],
45["emltg", 1, 1, "UNIFORM", 0.9, 2.1],
46["tmltg", 1.5, 1, "UNIFORM", 1, 2]]
The ‘use dist in hm?’ defines if the property will be history match (1) or the provided value in the second entry will be used (0).
Finally, we set if the permeabilities will be considered for the hm:
48# Coarser rock properties: name, use dist in hm?, coarsing approach (max or mean)
49rock = [["PERMX", 1, "max"],
50["PERMY", 0, "max"],
51["PERMZ", 0, "max"]]
If option 1 is selected, then the distributions are UNIFORM with interval values of [perm_min, perm_max], corresponding to the minimum and maximum values in each of the coarser cells.
Tip
By setting the “mode = ‘files’” in the toml configuration file, only the needed files to run a history matching using ERT are generated. Then one can inspect those files and do additional modifications before running the history matching by calling directly the ert executable. If you are not familiar with the format of the files to use ERT, visit the ERT documentation.